The G4 resolvase RHAU modulates mRNA translation and stability to sustain postnatal heart function and regeneration
- PMID: 33199370
- PMCID: PMC7948451
- DOI: 10.1074/jbc.RA120.014948
The G4 resolvase RHAU modulates mRNA translation and stability to sustain postnatal heart function and regeneration
Abstract
Post-transcriptional regulation of mRNA translation and stability is primarily achieved by RNA-binding proteins, which are of increasing importance for heart function. Furthermore, G-quadruplex (G4) and G4 resolvase activity are involved in a variety of biological processes. However, the role of G4 resolvase activity in heart function remains unknown. The present study aims to investigate the role of RNA helicase associated with adenylate- and uridylate-rich element (RHAU), an RNA-binding protein with G4 resolvase activity in postnatal heart function through deletion of Rhau in the cardiomyocytes of postnatal mice. RHAU-deficient mice displayed progressive pathological remodeling leading to heart failure and mortality and impaired neonatal heart regeneration. RHAU ablation reduced the protein levels but enhanced mRNA levels of Yap1 and Hexim1 that are important regulators for heart development and postnatal heart function. Furthermore, RHAU was found to associate with both the 5' and 3' UTRs of these genes to destabilize mRNA and enhance translation. Thus, we have demonstrated the important functions of RHAU in the dual regulation of mRNA translation and stability, which is vital for heart physiology.
Keywords: HEXIM1; RHAU; RNA-binding protein; YAP1; heart; mRNA; regeneration.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures
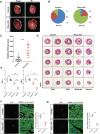
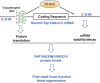
Similar articles
-
To unwind the biological knots: The DNA/RNA G-quadruplex resolvase RHAU (DHX36) in development and disease.Animal Model Exp Med. 2022 Dec;5(6):542-549. doi: 10.1002/ame2.12251. Epub 2022 Jul 4. Animal Model Exp Med. 2022. PMID: 35789129 Free PMC article. Review.
-
The G4 resolvase RHAU regulates ventricular trabeculation and compaction through transcriptional and post-transcriptional mechanisms.J Biol Chem. 2022 Jan;298(1):101449. doi: 10.1016/j.jbc.2021.101449. Epub 2021 Nov 25. J Biol Chem. 2022. PMID: 34838591 Free PMC article.
-
Post-transcriptional Regulation of Nkx2-5 by RHAU in Heart Development.Cell Rep. 2015 Oct 27;13(4):723-732. doi: 10.1016/j.celrep.2015.09.043. Epub 2015 Oct 17. Cell Rep. 2015. PMID: 26489465
-
A G-quadruplex DNA structure resolvase, RHAU, is essential for spermatogonia differentiation.Cell Death Dis. 2015 Jan 22;6(1):e1610. doi: 10.1038/cddis.2014.571. Cell Death Dis. 2015. PMID: 25611385 Free PMC article.
-
RHAU Peptides Specific for Parallel G-Quadruplexes: Potential Applications in Chemical Biology.Mol Biotechnol. 2023 Mar;65(3):291-299. doi: 10.1007/s12033-022-00552-7. Epub 2022 Aug 19. Mol Biotechnol. 2023. PMID: 35984625 Review.
Cited by
-
Study on the Interaction of a Peptide Targeting Specific G-Quadruplex Structures Based on Chromatographic Retention Behavior.Int J Mol Sci. 2023 Jan 11;24(2):1438. doi: 10.3390/ijms24021438. Int J Mol Sci. 2023. PMID: 36674950 Free PMC article.
-
Superfamily II helicases: the potential therapeutic target for cardiovascular diseases.Front Cardiovasc Med. 2023 Dec 13;10:1309491. doi: 10.3389/fcvm.2023.1309491. eCollection 2023. Front Cardiovasc Med. 2023. PMID: 38152606 Free PMC article. Review.
-
The G4 resolvase Dhx36 modulates cardiomyocyte differentiation and ventricular conduction system development.Nat Commun. 2024 Oct 4;15(1):8602. doi: 10.1038/s41467-024-52809-1. Nat Commun. 2024. PMID: 39366945 Free PMC article.
-
RNA-Binding Proteins as Critical Post-Transcriptional Regulators of Cardiac Regeneration.Int J Mol Sci. 2023 Jul 26;24(15):12004. doi: 10.3390/ijms241512004. Int J Mol Sci. 2023. PMID: 37569379 Free PMC article. Review.
-
To unwind the biological knots: The DNA/RNA G-quadruplex resolvase RHAU (DHX36) in development and disease.Animal Model Exp Med. 2022 Dec;5(6):542-549. doi: 10.1002/ame2.12251. Epub 2022 Jul 4. Animal Model Exp Med. 2022. PMID: 35789129 Free PMC article. Review.
References
-
- Krebs J.E., Lewin B., Goldstein E.S., Kilpatrick S.T. Jones & Bartlett Publishers, Burlington, MA; 2014. Lewin's Genes XI.
-
- Alberts B. Garland Science, New York, NY; 2017. Molecular Biology of the Cell.
-
- Lee T.I., Young R.A. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 2000;34:77–137. - PubMed
-
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. - PubMed
-
- Jaenisch R., Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33 Suppl:245–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

