Functional predication of differentially expressed circRNAs/lncRNAs in the prefrontal cortex of Nrf2-knockout mice
- PMID: 33714958
- PMCID: PMC8034947
- DOI: 10.18632/aging.202688
Functional predication of differentially expressed circRNAs/lncRNAs in the prefrontal cortex of Nrf2-knockout mice
Abstract
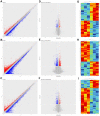
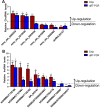
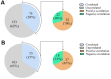
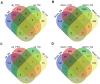
In the central nervous system, nuclear factor erythroid-2-related factor 2 (Nrf2) protects neurons from oxidant injury, thereby ameliorating neurodegeneration. We explored the key circular RNAs (circRNAs) and long non-coding RNAs (lncRNAs) involved in Nrf2-induced neuroprotection. We used microarrays to examine the circRNAs (DEcircRNAs), lncRNAs (DElncRNAs) and mRNAs (DEmRNAs) differentially expressed between Nrf2 (+/+) and Nrf2 (-/-) mice and identified DEcircRNA/DElncRNA-miRNA-DEmRNA interaction networks. In total, 197 DEcircRNAs, 685 DElncRNAs and 356 DEmRNAs were identified in prefrontal cortical tissues from Nrf2 (-/-) mice. The expression patterns of selected DEcircRNAs (except for mmu_circ_0003404) and DElncRNAs in qRT-PCR analyses were generally consistent with the microarray analysis results. Functional annotation of the DEmRNAs in the DEcircRNA/DElncRNA-miRNA-DEmRNA networks indicated that five non-coding RNAs (mmu_circ_0000233, ENSMUST00000204847, NONMMUT024778, NONMMUT132160 and NONMMUT132168) may contribute to Nrf2 activity, with the help of mmu_circ_0015035 and NONMMUT127961. The results also revealed that four non-coding RNAs (cicRNA.20127, mmu_circ_0012936, ENSMUST00000194077 and NONMMUT109267) may influence glutathione metabolism. Additionally, 44 DEcircRNAs and 7 DElncRNAs were found to possess coding potential. These findings provide clues to the molecular pathways through which Nrf2 protects neurons.
Keywords: Nrf2; circular RNA; long non-coding RNA.
Conflict of interest statement
Figures


Similar articles
-
The Differentially Expressed Circular RNAs in the Substantia Nigra and Corpus Striatum of Nrf2-Knockout Mice.Cell Physiol Biochem. 2018;50(3):936-951. doi: 10.1159/000494478. Epub 2018 Oct 24. Cell Physiol Biochem. 2018. PMID: 30355941
-
Differential Expression Profiles and Functional Prediction of Circular RNAs and Long Non-coding RNAs in the Hippocampus of Nrf2-Knockout Mice.Front Mol Neurosci. 2019 Aug 9;12:196. doi: 10.3389/fnmol.2019.00196. eCollection 2019. Front Mol Neurosci. 2019. PMID: 31447646 Free PMC article.
-
Identification of crucial noncoding RNAs and mRNAs in hypertrophic scars via RNA sequencing.FEBS Open Bio. 2021 Jun;11(6):1673-1684. doi: 10.1002/2211-5463.13167. Epub 2021 May 12. FEBS Open Bio. 2021. PMID: 33932142 Free PMC article.
-
Reconstruction and Analysis of the Differentially Expressed IncRNA-miRNA-mRNA Network Based on Competitive Endogenous RNA in Hepatocellular Carcinoma.Crit Rev Eukaryot Gene Expr. 2019;29(6):539-549. doi: 10.1615/CritRevEukaryotGeneExpr.2019028740. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422009 Review.
-
CircRNAs and LncRNAs in Osteoporosis.Differentiation. 2020 Nov-Dec;116:16-25. doi: 10.1016/j.diff.2020.10.002. Epub 2020 Oct 28. Differentiation. 2020. PMID: 33157509 Review.
Cited by
-
Role of Non-coding RNAs in Axon Regeneration after Peripheral Nerve Injury.Int J Biol Sci. 2022 May 9;18(8):3435-3446. doi: 10.7150/ijbs.70290. eCollection 2022. Int J Biol Sci. 2022. PMID: 35637962 Free PMC article. Review.
-
Response to comments on our article (Yin YL et al., Parasit Vectors, 10.1186/s13071-021-04739-w) by Yuqing Wang and colleagues.Parasit Vectors. 2021 Sep 21;14(1):484. doi: 10.1186/s13071-021-04996-9. Parasit Vectors. 2021. PMID: 34548103 Free PMC article.
References
-
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997; 236:313–22. 10.1006/bbrc.1997.6943 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

