Enhancing Ristomycin A Production by Overexpression of ParB-Like StrR Family Regulators Controlling the Biosynthesis Genes
- PMID: 34505824
- PMCID: PMC8432530
- DOI: 10.1128/AEM.01066-21
Enhancing Ristomycin A Production by Overexpression of ParB-Like StrR Family Regulators Controlling the Biosynthesis Genes
Abstract
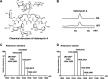
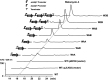
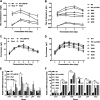
Amycolatopsis sp. strain TNS106 harbors a ristomycin-biosynthetic gene cluster (asr) in its genome and produces ristomycin A. Deletion of the sole cluster-situated StrR family regulatory gene, asrR, abolished ristomycin A production and the transcription of the asr genes orf5 to orf39. The ristomycin A fermentation titer in Amycolatopsis sp. strain TNS106 was dramatically improved by overexpression of asrR and a heterologous StrR family regulatory gene, bbr, from the balhimycin-biosynthetic gene cluster (BGC) utilizing strong promoters and multiple gene copies. Ristomycin A production was improved by approximately 60-fold, resulting in a fermentation titer of 4.01 g/liter in flask culture, in one of the engineered strains. Overexpression of AsrR and Bbr upregulated transcription of tested asr biosynthetic genes, indicating that these asr genes were positively regulated by AsrR and Bbr. However, only the promoter region of the asrR operon and the intergenic region upstream of orf12 were bound by AsrR and Bbr in gel retardation assays, suggesting that AsrR and Bbr directly regulated the asrR operon and probably orf12 to orf14 but no other asr biosynthetic genes. Further assays with synthetic short probes showed that AsrR and Bbr specifically bound not only probes containing the canonical inverted repeats but also a probe with only one 7-bp element of the inverted repeats in its native context. AsrR and Bbr have an N-terminal ParB-like domain and a central winged helix-turn-helix DNA-binding domain. Site-directed mutations indicated that the N-terminal ParB-like domain was involved in activation of ristomycin A biosynthesis and did not affect the DNA-binding activity of AsrR and Bbr. IMPORTANCE This study showed that overexpression of either a native StrR family regulator (AsrR) or a heterologous StrR family regulator (Bbr) dramatically improved ristomycin A production by increasing the transcription of biosynthetic genes directly or indirectly. The conserved ParB-like domain of AsrR and Bbr was demonstrated to be involved in the regulation of asr BGC expression. These findings provide new insights into the mechanism of StrR family regulators in the regulation of glycopeptide antibiotic biosynthesis. Furthermore, the regulator overexpression plasmids constructed in this study could serve as valuable tools for strain improvement and genome mining for new glycopeptide antibiotics. In addition, ristomycin A is a type III glycopeptide antibiotic clinically used as a diagnostic reagent due to its side effects. The overproduction strains engineered in this study are ideal materials for industrial production of ristomycin A.
Keywords: Amycolatopsis; N-terminal ParB-like domain; StrR family regulator; antibiotic titer improvement; glycopeptide antibiotics; ristomycin.
Figures




Similar articles
-
The border sequence of the balhimycin biosynthesis gene cluster from Amycolatopsis balhimycina contains bbr, encoding a StrR-like pathway-specific regulator.J Mol Microbiol Biotechnol. 2007;13(1-3):76-88. doi: 10.1159/000103599. J Mol Microbiol Biotechnol. 2007. PMID: 17693715
-
Overproduction of Ristomycin A by activation of a silent gene cluster in Amycolatopsis japonicum MG417-CF17.Antimicrob Agents Chemother. 2014 Oct;58(10):6185-96. doi: 10.1128/AAC.03512-14. Epub 2014 Aug 11. Antimicrob Agents Chemother. 2014. PMID: 25114137 Free PMC article.
-
Regulation of teicoplanin biosynthesis: refining the roles of tei cluster-situated regulatory genes.Appl Microbiol Biotechnol. 2019 May;103(10):4089-4102. doi: 10.1007/s00253-019-09789-w. Epub 2019 Apr 1. Appl Microbiol Biotechnol. 2019. PMID: 30937499
-
Metabolic engineering of the shikimate pathway in Amycolatopsis strains for optimized glycopeptide antibiotic production.Metab Eng. 2023 Jul;78:84-92. doi: 10.1016/j.ymben.2023.05.005. Epub 2023 May 25. Metab Eng. 2023. PMID: 37244369
-
NovG, a DNA-binding protein acting as a positive regulator of novobiocin biosynthesis.Microbiology (Reading). 2005 Jun;151(Pt 6):1949-1961. doi: 10.1099/mic.0.27669-0. Microbiology (Reading). 2005. PMID: 15942002
Cited by
-
The Impact of Heterologous Regulatory Genes from Lipodepsipeptide Biosynthetic Gene Clusters on the Production of Teicoplanin and A40926.Antibiotics (Basel). 2024 Jan 24;13(2):115. doi: 10.3390/antibiotics13020115. Antibiotics (Basel). 2024. PMID: 38391501 Free PMC article.
-
Genetics Behind the Glycosylation Patterns in the Biosynthesis of Dalbaheptides.Front Chem. 2022 Mar 24;10:858708. doi: 10.3389/fchem.2022.858708. eCollection 2022. Front Chem. 2022. PMID: 35402387 Free PMC article. Review.
-
Development of Integrated Vectors with Strong Constitutive Promoters for High-Yield Antibiotic Production in Mangrove-Derived Streptomyces.Mar Drugs. 2024 Feb 18;22(2):94. doi: 10.3390/md22020094. Mar Drugs. 2024. PMID: 38393065 Free PMC article.
-
Three new LmbU targets outside lmb cluster inhibit lincomycin biosynthesis in Streptomyces lincolnensis.Microb Cell Fact. 2024 Jan 3;23(1):3. doi: 10.1186/s12934-023-02284-y. Microb Cell Fact. 2024. PMID: 38172890 Free PMC article.
-
Gram-Level Production of Balanol through Regulatory Pathway and Medium Optimization in Herb Fungus Tolypocladium ophioglossoides.J Fungi (Basel). 2022 May 16;8(5):510. doi: 10.3390/jof8050510. J Fungi (Basel). 2022. PMID: 35628765 Free PMC article.
References
-
- Pelzer S, Süssmuth R, Heckmann D, Recktenwald J, Huber P, Jung G, Wohlleben W. 1999. Identification and analysis of the balhimycin biosynthetic gene cluster and its use for manipulating glycopeptide biosynthesis in Amycolatopsis mediterranei DSM5908. Antimicrob Agents Chemother 43:1565–1573. 10.1128/AAC.43.7.1565. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

