Lipotoxic hepatocyte-derived exosomal miR-1297 promotes hepatic stellate cell activation through the PTEN signaling pathway in metabolic-associated fatty liver disease
- PMID: 33911465
- PMCID: PMC8047533
- DOI: 10.3748/wjg.v27.i14.1419
Lipotoxic hepatocyte-derived exosomal miR-1297 promotes hepatic stellate cell activation through the PTEN signaling pathway in metabolic-associated fatty liver disease
Abstract
Background: Exosomes play an important role in metabolic-associated fatty liver disease (MAFLD), but the mechanism by which exosomes participate in MAFLD still remain unclear.
Aim: To figure out the function of lipotoxic exosomal miR-1297 in MAFLD.
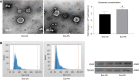
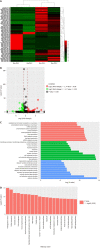
Methods: MicroRNA sequencing was used to detect differentially expressed miRNAs (DE-miR) in lipotoxic exosomes derived from primary hepatocytes. Bioinformatic tools were applied to analyze the target genes and pathways regulated by the DE-miRs. Quantitative real-time PCR (qPCR) was conducted for the verification of DE-miRs. qPCR, western blot, immunofluorescence staining and ethynyl-20-deoxyuridine assay were used to evaluate the function of lipotoxic exosomal miR-1297 on hepatic stellate cells (LX2 cells). A luciferase reporter experiment was performed to confirm the relationship of miR-1297 and its target gene PTEN.
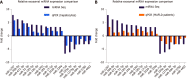
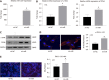
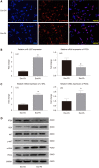
Results: MicroRNA sequencing revealed that there were 61 exosomal DE-miRs (P < 0.05) with a fold-change > 2 from palmitic acid treated primary hepatocytes compared with the vehicle control group. miR-1297 was the most highly upregulated according to the microRNA sequencing. Bioinformatic tools showed a variety of target genes and pathways regulated by these DE-miRs were related to liver fibrosis. miR-1297 was overexpressed in exosomes derived from lipotoxic hepatocytes by qPCR. Fibrosis promoting genes (α-SMA, PCNA) were altered in LX2 cells after miR-1297 overexpression or miR-1297-rich lipotoxic exosome incubation via qPCR and western blot analysis. Immunofluorescence staining and ethynyl-20-deoxyuridine staining demonstrated that the activation and proliferation of LX2 cells were also promoted after the above treatment. PTEN was found to be the target gene of miR-1297 and knocking down PTEN contributed to the activation and proliferation of LX2 cells via modulating the PI3K/AKT signaling pathway.
Conclusion: miR-1297 was overexpressed in exosomes derived from lipotoxic hepatocytes. The lipotoxic hepatocyte-derived exosomal miR-1297 could promote the activation and proliferation of hepatic stellate cells through the PTEN/PI3K/AKT signaling pathway, accelerating the progression of MAFLD.
Keywords: Exosome; Hepatic stellate cell; Liver fibrosis; Metabolic-associated fatty liver disease; PTEN; miRNA-1297.
©The Author(s) 2021. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: The authors hereby declare that no conflict of interest exists.
Figures

Similar articles
-
Lipotoxic Hepatocyte-Derived Exosomal MicroRNA 192-5p Activates Macrophages Through Rictor/Akt/Forkhead Box Transcription Factor O1 Signaling in Nonalcoholic Fatty Liver Disease.Hepatology. 2020 Aug;72(2):454-469. doi: 10.1002/hep.31050. Epub 2020 May 16. Hepatology. 2020. PMID: 31782176 Free PMC article.
-
HepG2.2.15-derived exosomes facilitate the activation and fibrosis of hepatic stellate cells.World J Gastroenterol. 2024 May 21;30(19):2553-2563. doi: 10.3748/wjg.v30.i19.2553. World J Gastroenterol. 2024. PMID: 38817658 Free PMC article.
-
Hepatocyte-derived exosomal miR-27a activateshepatic stellate cells through the inhibitionof PINK1-mediated mitophagy in MAFLD.Mol Ther Nucleic Acids. 2021 Nov 3;26:1241-1254. doi: 10.1016/j.omtn.2021.10.022. eCollection 2021 Dec 3. Mol Ther Nucleic Acids. 2021. PMID: 34853724 Free PMC article.
-
Exosomal microRNAs as diagnostic and therapeutic biomarkers in non-malignant liver diseases.Arch Pharm Res. 2021 Jun;44(6):574-587. doi: 10.1007/s12272-021-01338-2. Epub 2021 Jun 24. Arch Pharm Res. 2021. PMID: 34165701 Free PMC article. Review.
-
Recent advances in the role of exosomes in liver fibrosis.J Gastroenterol Hepatol. 2023 Jul;38(7):1083-1088. doi: 10.1111/jgh.16203. Epub 2023 Apr 28. J Gastroenterol Hepatol. 2023. PMID: 37114594 Review.
Cited by
-
Increased Hepatic ATG7 mRNA and ATG7 Protein Expression in Nonalcoholic Steatohepatitis Associated with Obesity.Int J Mol Sci. 2023 Jan 10;24(2):1324. doi: 10.3390/ijms24021324. Int J Mol Sci. 2023. PMID: 36674839 Free PMC article.
-
Role of exosomes in the development of the immune microenvironment in hepatocellular carcinoma.Front Immunol. 2023 Jun 29;14:1200201. doi: 10.3389/fimmu.2023.1200201. eCollection 2023. Front Immunol. 2023. PMID: 37457718 Free PMC article. Review.
-
Nucleic acids and proteins carried by exosomes from various sources: Potential role in liver diseases.Front Physiol. 2022 Sep 23;13:957036. doi: 10.3389/fphys.2022.957036. eCollection 2022. Front Physiol. 2022. PMID: 36213232 Free PMC article. Review.
-
Autophagy Dysregulation in Metabolic Associated Fatty Liver Disease: A New Therapeutic Target.Int J Mol Sci. 2022 Sep 2;23(17):10055. doi: 10.3390/ijms231710055. Int J Mol Sci. 2022. PMID: 36077452 Free PMC article. Review.
-
The regulatory role and mechanism of exosomes in hepatic fibrosis.Front Pharmacol. 2023 Dec 1;14:1284742. doi: 10.3389/fphar.2023.1284742. eCollection 2023. Front Pharmacol. 2023. PMID: 38108065 Free PMC article. Review.
References
-
- Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, Bugianesi E. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11–20. - PubMed
-
- Eslam M, Sanyal AJ, George J International Consensus Panel. MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology 2020; 158: 1999-2014. :e1. - PubMed
-
- Fan JG, Kim SU, Wong VW. New trends on obesity and NAFLD in Asia. J Hepatol. 2017;67:862–873. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

