Suppressed inflammation in obese children induced by a high-fiber diet is associated with the attenuation of gut microbial virulence factor genes
- PMID: 34233588
- PMCID: PMC8274444
- DOI: 10.1080/21505594.2021.1948252
Suppressed inflammation in obese children induced by a high-fiber diet is associated with the attenuation of gut microbial virulence factor genes
Abstract
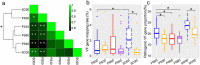
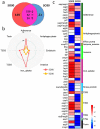
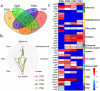
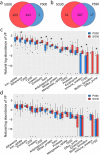
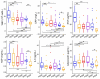
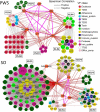
In our previous study, a gut microbiota-targeted dietary intervention with a high-fiber diet improved the immune status of both genetically obese (Prader-Willi Syndrome, PWS) and simple obese (SO) children. However, PWS children had higher inflammation levels than SO children throughout the trial, the gut microbiota of the two cohorts was similar. As some virulence factors (VFs) produced by the gut microbiota play a role in triggering host inflammation, this study compared the characteristics and changes of gut microbial VF genes of the two cohorts before and after the intervention using a fecal metagenomic dataset. We found that in both cohorts, the high-fiber diet reduced the abundance of VF, and particularly pathogen-specific, genes. The composition of VF genes was also modulated, especially for offensive and defensive VF genes. Furthermore, genes belonging to invasion, T3SS (type III secretion system), and adherence classes were suppressed. Co-occurrence network analysis detected VF gene clusters closely related to host inflammation in each cohort. Though these cohort-specific clusters varied in VF gene combinations and cascade reactions affecting inflammation, they mainly contained VFs belonging to iron uptake, T3SS, and invasion classes. The PWS group had a lower abundance of VF genes before the trial, which suggested that other factors could also be responsible for the increased inflammation in this cohort. This study provides insight into the modulation of VF gene structure in the gut microbiota by a high-fiber diet, with respect to reduced inflammation in obese children, and differences in VF genes between these two cohorts.
Keywords: Virulence factor; gut microbiota; high-fiber diet; inflammation; metagenomic; obesity; prader-willi syndrome.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
Similar articles
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
-
Mortality impact, risks, and benefits of general population screening for ovarian cancer: the UKCTOCS randomised controlled trial.Health Technol Assess. 2023 May 11:1-81. doi: 10.3310/BHBR5832. Online ahead of print. Health Technol Assess. 2023. PMID: 37183782 Free PMC article.
-
Dacryocystorhinostomy.2023 Aug 7. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2023 Aug 7. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 32496731 Free Books & Documents.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article. Review.
Cited by
-
Metagenome-Scale Metabolic Network Suggests Folate Produced by Bifidobacterium longum Might Contribute to High-Fiber-Diet-Induced Weight Loss in a Prader-Willi Syndrome Child.Microorganisms. 2021 Dec 1;9(12):2493. doi: 10.3390/microorganisms9122493. Microorganisms. 2021. PMID: 34946095 Free PMC article.
-
Type III Secretion System in Intestinal Pathogens and Metabolic Diseases.J Diabetes Res. 2024 Nov 6;2024:4864639. doi: 10.1155/2024/4864639. eCollection 2024. J Diabetes Res. 2024. PMID: 39544522 Free PMC article. Review.
-
Associations between Oxidant/Antioxidant Status and Circulating Adipokines in Non-Obese Children with Prader-Willi Syndrome.Antioxidants (Basel). 2023 Apr 13;12(4):927. doi: 10.3390/antiox12040927. Antioxidants (Basel). 2023. PMID: 37107302 Free PMC article.
-
Dietary Inflammatory Potential in Pediatric Diseases: A Narrative Review.Nutrients. 2023 Dec 13;15(24):5095. doi: 10.3390/nu15245095. Nutrients. 2023. PMID: 38140353 Free PMC article. Review.
-
Bacteroides acidifaciens in the gut plays a protective role against CD95-mediated liver injury.Gut Microbes. 2022 Jan-Dec;14(1):2027853. doi: 10.1080/19490976.2022.2027853. Gut Microbes. 2022. PMID: 35129072 Free PMC article.
References
-
- Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6(1):6528. - PubMed
-
- Wang XK, Xu XQ, Xia Y.. Further analysis reveals new gut microbiome markers of type 2 diabetes mellitus. Anton Leeuw Int J G. 2017;110(3):445–453. - PubMed
-
- Kamada N, Seo SU, Chen GY, et al. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–335. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
