DDX56 modulates post-transcriptional Wnt signaling through miRNAs and is associated with early recurrence in squamous cell lung carcinoma
- PMID: 34446021
- PMCID: PMC8393456
- DOI: 10.1186/s12943-021-01403-w
DDX56 modulates post-transcriptional Wnt signaling through miRNAs and is associated with early recurrence in squamous cell lung carcinoma
Abstract
Background: Early recurrence is a major obstacle to prolonged postoperative survival in squamous cell lung carcinoma (SqCLC). The molecular mechanisms underlying early SqCLC recurrence remain unclear, and effective prognostic biomarkers for predicting early recurrence are needed.
Methods: We analyzed primary tumor samples of 20 SqCLC patients using quantitative proteomics to identify differentially-expressed proteins in patients who experienced early versus late disease recurrence. The expression and prognostic significance of DDX56 was evaluated using a SqCLC tumor tissue microarray and further verified using different online databases. We performed in vitro and in vivo experiments to obtain detailed molecular insight into the functional role of DDX56 in SqCLC.
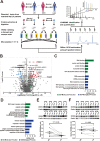
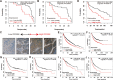
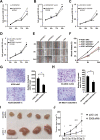
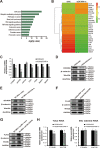
Results: We found that DDX56 exhibited increased expression in tumors of patients who experienced early versus late disease recurrence. Increased DDX56 expression in SqCLC tumors was subsequently confirmed as an independent prognostic factor of poor recurrence-free survival in independent SqCLC cohorts. Functionally, DDX56 promotes SqCLC cell growth and migration in vitro, and xenograft tumor progression in vivo. Mechanistically, DDX56 post-transcriptionally promotes expression of multiple Wnt signaling pathway-related genes, including CTNNB1, WNT2B, and represses a subset of miRNAs, including miR-378a-3p, a known suppressor of Wnt signaling. Detailed analysis revealed that DDX56 facilitated degradation of primary miR-378a, leading to down-regulation of mature miR-378a-3p and thus derepression of the target gene WNT2B.
Conclusion: We identified DDX56 as a novel independent prognostic biomarker that exerts its oncogenic effects through miRNA-mediated post-transcriptional regulation of Wnt signaling genes to promote early SqCLC recurrence. DDX56 may assist in identifying SqCLC patients at increased risk of early recurrence and who could benefit from Wnt signaling-targeted therapies.
Keywords: DDX56; Squamous cell lung cancer; Wnt signaling pathway; miRNA.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
The authors confirm that there is no conflict of interest related to this manuscript.
Figures

Similar articles
-
miR-140-3p functions as a tumor suppressor in squamous cell lung cancer by regulating BRD9.Cancer Lett. 2019 Apr 1;446:81-89. doi: 10.1016/j.canlet.2019.01.007. Epub 2019 Jan 17. Cancer Lett. 2019. PMID: 30660651
-
Expression of fibroblast growth factor receptor 1, fibroblast growth factor 2, phosphatidyl inositol 3 phosphate kinase and their clinical and prognostic significance in early and advanced stage of squamous cell carcinoma of the lung.Int J Clin Exp Pathol. 2015 Sep 1;8(9):9760-71. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 26617686 Free PMC article.
-
DEAD-box helicase 56 functions as an oncogene promote cell proliferation and invasion in gastric cancer via the FOXO1/p21 Cip1/c-Myc signaling pathway.Bioengineered. 2022 May;13(5):13970-13985. doi: 10.1080/21655979.2022.2084235. Bioengineered. 2022. PMID: 35723050 Free PMC article.
-
DDX56 transcriptionally activates MIST1 to facilitate tumorigenesis of HCC through PTEN-AKT signaling.Theranostics. 2022 Aug 15;12(14):6069-6087. doi: 10.7150/thno.72471. eCollection 2022. Theranostics. 2022. PMID: 36168636 Free PMC article.
-
[Research status on molecular targeted therapy for squamous-cell lung cancer].Zhongguo Fei Ai Za Zhi. 2014 Aug 20;17(8):618-24. doi: 10.3779/j.issn.1009-3419.2014.08.07. Zhongguo Fei Ai Za Zhi. 2014. PMID: 25130969 Free PMC article. Review. Chinese.
Cited by
-
DDX56 promotes EMT and cancer stemness via MELK-FOXM1 axis in hepatocellular carcinoma.iScience. 2024 Apr 29;27(6):109827. doi: 10.1016/j.isci.2024.109827. eCollection 2024 Jun 21. iScience. 2024. PMID: 38827395 Free PMC article.
-
Role and therapeutic potential of DEAD-box RNA helicase family in colorectal cancer.Front Oncol. 2023 Oct 27;13:1278282. doi: 10.3389/fonc.2023.1278282. eCollection 2023. Front Oncol. 2023. PMID: 38023215 Free PMC article. Review.
-
Upregulation of Wnt2b exerts neuroprotective effect by alleviating mitochondrial dysfunction in Alzheimer's disease.CNS Neurosci Ther. 2023 Jul;29(7):1805-1816. doi: 10.1111/cns.14139. Epub 2023 Feb 27. CNS Neurosci Ther. 2023. PMID: 36852442 Free PMC article.
-
Noncoding RNAs-mediated overexpression of KIF14 is associated with tumor immune infiltration and unfavorable prognosis in lung adenocarcinoma.Aging (Albany NY). 2022 Oct 11;14(19):8013-8031. doi: 10.18632/aging.204332. Epub 2022 Oct 11. Aging (Albany NY). 2022. PMID: 36227151 Free PMC article.
-
Pan-cancer analysis identifies DDX56 as a prognostic biomarker associated with immune infiltration and drug sensitivity.Front Genet. 2022 Dec 7;13:1004467. doi: 10.3389/fgene.2022.1004467. eCollection 2022. Front Genet. 2022. PMID: 36568395 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

