USP12 downregulation orchestrates a protumourigenic microenvironment and enhances lung tumour resistance to PD-1 blockade
- PMID: 34381028
- PMCID: PMC8357983
- DOI: 10.1038/s41467-021-25032-5
USP12 downregulation orchestrates a protumourigenic microenvironment and enhances lung tumour resistance to PD-1 blockade
Abstract
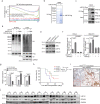
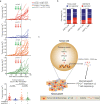
Oncogenic activation of KRAS and its surrogates is essential for tumour cell proliferation and survival, as well as for the development of protumourigenic microenvironments. Here, we show that the deubiquitinase USP12 is commonly downregulated in the KrasG12D-driven mouse lung tumour and human non-small cell lung cancer owing to the activation of AKT-mTOR signalling. Downregulation of USP12 promotes lung tumour growth and fosters an immunosuppressive microenvironment with increased macrophage recruitment, hypervascularization, and reduced T cell activation. Mechanistically, USP12 downregulation creates a tumour-promoting secretome resulting from insufficient PPM1B deubiquitination that causes NF-κB hyperactivation in tumour cells. Furthermore, USP12 inhibition desensitizes mouse lung tumour cells to anti-PD-1 immunotherapy. Thus, our findings propose a critical component downstream of the oncogenic signalling pathways in the modulation of tumour-immune cell interactions and tumour response to immune checkpoint blockade therapy.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Immune reprogramming via PD-1 inhibition enhances early-stage lung cancer survival.JCI Insight. 2018 Jul 12;3(13):e96836. doi: 10.1172/jci.insight.96836. JCI Insight. 2018. PMID: 29997286 Free PMC article.
-
Synergic effect of PD-1 blockade and endostar on the PI3K/AKT/mTOR-mediated autophagy and angiogenesis in Lewis lung carcinoma mouse model.Biomed Pharmacother. 2020 May;125:109746. doi: 10.1016/j.biopha.2019.109746. Epub 2020 Feb 25. Biomed Pharmacother. 2020. PMID: 32106386
-
The progress and confusion of anti-PD1/PD-L1 immunotherapy for patients with advanced non-small cell lung cancer.Int Immunopharmacol. 2020 Mar;80:106247. doi: 10.1016/j.intimp.2020.106247. Epub 2020 Jan 31. Int Immunopharmacol. 2020. PMID: 32007710 Review.
-
ILT4 inhibition prevents TAM- and dysfunctional T cell-mediated immunosuppression and enhances the efficacy of anti-PD-L1 therapy in NSCLC with EGFR activation.Theranostics. 2021 Jan 19;11(7):3392-3416. doi: 10.7150/thno.52435. eCollection 2021. Theranostics. 2021. PMID: 33537094 Free PMC article.
-
Immunotherapy in NSCLC patients with brain metastases. Understanding brain tumor microenvironment and dissecting outcomes from immune checkpoint blockade in the clinic.Cancer Treat Rev. 2020 Sep;89:102067. doi: 10.1016/j.ctrv.2020.102067. Epub 2020 Jul 7. Cancer Treat Rev. 2020. PMID: 32682248 Review.
Cited by
-
Drug resistance mechanisms and treatment strategies mediated by Ubiquitin-Specific Proteases (USPs) in cancers: new directions and therapeutic options.Mol Cancer. 2024 May 3;23(1):88. doi: 10.1186/s12943-024-02005-y. Mol Cancer. 2024. PMID: 38702734 Free PMC article. Review.
-
USP12 promotes antiviral responses by deubiquitinating and stabilizing IFI16.PLoS Pathog. 2023 Jul 6;19(7):e1011480. doi: 10.1371/journal.ppat.1011480. eCollection 2023 Jul. PLoS Pathog. 2023. PMID: 37410794 Free PMC article.
-
Ubiquitination and deubiquitination in cancer: from mechanisms to novel therapeutic approaches.Mol Cancer. 2024 Jul 25;23(1):148. doi: 10.1186/s12943-024-02046-3. Mol Cancer. 2024. PMID: 39048965 Free PMC article. Review.
-
Identification of key biomarkers based on the proliferation of secondary hyperparathyroidism by bioinformatics analysis and machine learning.PeerJ. 2023 Jul 10;11:e15633. doi: 10.7717/peerj.15633. eCollection 2023. PeerJ. 2023. PMID: 37456892 Free PMC article.
-
Exploiting E3 ubiquitin ligases to reeducate the tumor microenvironment for cancer therapy.Exp Hematol Oncol. 2023 Mar 30;12(1):34. doi: 10.1186/s40164-023-00394-2. Exp Hematol Oncol. 2023. PMID: 36998063 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

