Divergent RNA viruses in Macrophomina phaseolina exhibit potential as virocontrol agents
- PMID: 33505706
- PMCID: PMC7816680
- DOI: 10.1093/ve/veaa095
Divergent RNA viruses in Macrophomina phaseolina exhibit potential as virocontrol agents
Abstract
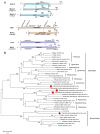
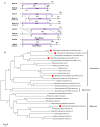
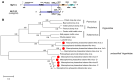
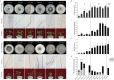
Macrophomina phaseolina is an important necrotrophic phytopathogenic fungus and cause extensive damage in many oilseed crops. Twelve M.phaseolina isolates with diverse biological phenotypes were selected for a high-throughput sequencing-based metatranscriptomic and bioinformatics analysis to identify viruses infecting M.phaseolina. The analysis identified 40 partial or nearly complete viral genome segments, 31 of which were novel viruses. Among these viral sequences, 43% of the viral genomes were double-stranded RNA (dsRNA), 47% were positive single-stranded RNA (ssRNA+), and the remaining 10% were negative sense-stranded RNA (ssRNA-). The 40 viruses showed affinity to 13 distinct viral lineages, including Bunyavirales (four viruses), Totiviridae (three viruses), Chrysoviridae (five viruses), Partitiviridae (four viruses), Hypoviridae (one virus), Endornaviridae (two viruses), Tombusviridae (three viruses), Narnaviridae (one virus), Potyviridae (one virus), Bromoviridae (one virus), Virgaviridae (six viruses), 'Fusagraviridae' (five viruses), and Ourmiavirus (four viruses). Two viruses are closely related to two families, Potyviridae and Bromoviridae, which previously contained no mycovirus species. Moreover, nine novel viruses associated with M.phaseolina were identified in the family Totiviridae, Endornaviridae, and Partitiviridae. Coinfection with multiple viruses is prevalent in M.phaseolina, with each isolate harboring different numbers of viruses, ranging from three to eighteen. Furthermore, the effects of the viruses on the fungal host were analyzed according to the biological characteristics of each isolate. The results suggested that M.phaseolina hypovirus 2, M.phaseolina fusagravirus virus 1-5 (MpFV1-5), M.phaseolina endornavirus 1-2 (MpEV1-2), M.phaseolina ourmia-like virus 1-3 (MpOLV1-3), M.phaseolina mitovirus 4 (MpMV4), and M.phaseolina mycobunyavirus 1-4 (MpMBV1-4) were only detected in hypovirulent isolates. Those viruses associated with hypovirulence might be used as biological control agents as an environmentally friendly alternative to chemical fungicides. These findings considerably expand our understanding of mycoviruses in M.phaseolina and unvailed the presence of a huge difference among viruses in isolates from different hosts in distant geographical regions. Together, the present study provides new knowledge about viral evolution and fungus-virus coevolution.
Keywords: Macrophomina phaseolina; coinfection; diversity; hypovirulence; metatranscriptomic; mycovirus.
© The Author(s) 2020. Published by Oxford University Press.
Figures





Similar articles
-
Identification of Diverse Mycoviruses through Metatranscriptomics Characterization of the Viromes of Five Major Fungal Plant Pathogens.J Virol. 2016 Jul 11;90(15):6846-6863. doi: 10.1128/JVI.00357-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27194764 Free PMC article.
-
A novel double-stranded RNA mycovirus that infects Macrophomina phaseolina.Arch Virol. 2019 Sep;164(9):2411-2416. doi: 10.1007/s00705-019-04334-6. Epub 2019 Jun 28. Arch Virol. 2019. PMID: 31254049
-
Co-infection of a hypovirulent isolate of Sclerotinia sclerotiorum with a new botybirnavirus and a strain of a mitovirus.Virol J. 2016 Jun 6;13:92. doi: 10.1186/s12985-016-0550-2. Virol J. 2016. PMID: 27267756 Free PMC article.
-
Viruses Infecting the Plant Pathogenic Fungus Rhizoctonia solani.Viruses. 2019 Nov 30;11(12):1113. doi: 10.3390/v11121113. Viruses. 2019. PMID: 31801308 Free PMC article. Review.
-
[Diverse double-stranded RNA viruses infecting fungi].Uirusu. 2014;64(2):225-38. doi: 10.2222/jsv.64.225. Uirusu. 2014. PMID: 26437844 Review. Japanese.
Cited by
-
Fungal Viruses Unveiled: A Comprehensive Review of Mycoviruses.Viruses. 2023 May 19;15(5):1202. doi: 10.3390/v15051202. Viruses. 2023. PMID: 37243288 Free PMC article. Review.
-
Four Novel Mycoviruses from the Hypovirulent Botrytis cinerea SZ-2-3y Isolate from Paris polyphylla: Molecular Characterisation and Mitoviral Sequence Transboundary Entry into Plants.Viruses. 2022 Jan 14;14(1):151. doi: 10.3390/v14010151. Viruses. 2022. PMID: 35062353 Free PMC article.
-
Novel and diverse mycoviruses co-infecting a single strain of the phytopathogenic fungus Alternaria dianthicola.Front Cell Infect Microbiol. 2022 Sep 27;12:980970. doi: 10.3389/fcimb.2022.980970. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36237429 Free PMC article.
-
Characterization of the RNA Mycovirome Associated with Grapevine Fungal Pathogens: Analysis of Mycovirus Distribution and Their Genetic Variability within a Collection of Botryosphaeriaceae Isolates.Viruses. 2024 Mar 1;16(3):392. doi: 10.3390/v16030392. Viruses. 2024. PMID: 38543758 Free PMC article.
-
Identification and characterization of mycoviruses in transcriptomes from the fungal family ceratocystidaceae.Virus Genes. 2024 Dec;60(6):696-710. doi: 10.1007/s11262-024-02112-4. Epub 2024 Oct 8. Virus Genes. 2024. PMID: 39378002 Free PMC article.
References
-
- Adams M. J., Antoniw J. F., Kreuze J. (2009) ‘ Virgaviridae: A New Family of Rod-Shaped Plant Viruses’, Archives of Virology, 154: 1967–72. - PubMed
-
- Adams Z. (2012) ‘Family Potyviridae’, in King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J. (eds.) Virus Taxonomy: Classification and Nomenclature of Viruses, Ninth Report of the International Committee on Taxonomy of Viruses, pp.1069–89. Waltham, MA, USA.
-
- Aiewsakun P., Katzourakis A. (2015) ‘ Endogenous Viruses: Connecting Recent and Ancient Viral Evolution’, Virology, 479–480: 26–37. - PubMed
-
- Ammon V., Wyllie T. D., Brown M. F. (1974) ‘ An Ultrastructural Investigation of Pathological Alterations Induced by Macrophomina Phaseolina (Tassi) Goid in Seedlings of Soybean, Glycine max (L.) Merrill’, Physiological Plant Pathology, 4: 1–4.
LinkOut - more resources
Full Text Sources

