Lnc-GAN1 expression is associated with good survival and suppresses tumor progression by sponging mir-26a-5p to activate PTEN signaling in non-small cell lung cancer
- PMID: 33407724
- PMCID: PMC7786923
- DOI: 10.1186/s13046-020-01819-0
Lnc-GAN1 expression is associated with good survival and suppresses tumor progression by sponging mir-26a-5p to activate PTEN signaling in non-small cell lung cancer
Abstract
Background: Long non-coding RNAs (lncRNAs) play vital roles in the development and progression of non-small-cell lung cancer (NSCLC); however, the role of most lncRNAs in NSCLC remains unknown. This study explored the clinical significance, biological function and underlying mechanism of lnc-GAN1 in NSCLC.
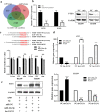
Methods: With a custom lncRNA microarray we found that lnc-GAN1 is markedly downregulated in NSCLC tissues. Then lnc-GAN1 expression level was measured using qRT-PCR in NSCLC tissues and cell lines. Survival was assessed using the Kaplan-Meier method. The biological functions of lnc-GAN1 in lung cancer cells were evaluated in vitro and in vivo. RNA fluorescence in situ hybridization and subcellular localization assays revealed the subcellular distribution of lnc-GAN1 in cells. Bioinformatic analysis was adopted to predict miRNAs and signaling pathways regulated by lnc-GAN1. RNA immunoprecipitation and Dual-luciferase reporter assays were used to assess the interaction between lnc-GAN1 and miR-26a-5p in lung cancer cells.
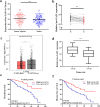
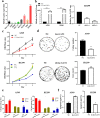
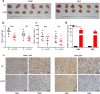
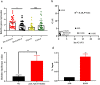
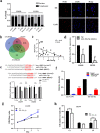
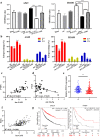
Results: lnc-GAN1 is downregulated in HCC tissues and associated with larger tumor size and poor overall survival and disease-free survival; its ectopic expression suppresses cell proliferation, colony formation, and cell cycle progression and induces apoptosis in NSCLC cells; it also inhibits tumor growth in the NSCLC xenograft model. We further proved that lnc-GAN1 is localized in cytoplasm and transcribed independently from its parental gene GAN. Mechanistically, lnc-GAN1 acts as a sponge for miR-26a-5p by two seed sequences, and the two non-coding RNAs have a negative relationship in NSCLC tissues; we further prove that PTEN is a direct target of miR-26a-5p and lnc-GAN1 inhibits cell cycle signaling pathway by activating PTEN, whose expression level correlated negatively with miR-26a-5p level but positively with lnc-GAN1 level in NSCLC samples.
Conclusions: Lnc-GAN1 is downregulated and associated with poor survival of NSCLC patients, and mechanistically acts as a tumor suppressor via sponging and inhibiting miR-26a-5p to upregulate PTEN. This study provides a potential prognostic biomarker and treatment target for NSCLC.
Keywords: Lnc-GAN1; Lung cancer; PTEN; miR-26a-5p.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
The Role of lncRNA-miR-26a-mRNA Network in Cancer Progression and Treatment.Biochem Genet. 2024 Jun;62(3):1443-1461. doi: 10.1007/s10528-023-10475-w. Epub 2023 Sep 20. Biochem Genet. 2024. PMID: 37730965 Review.
-
The Lnc LINC00461/miR-30a-5p facilitates progression and malignancy in non-small cell lung cancer via regulating ZEB2.Cell Cycle. 2020 Apr;19(7):825-836. doi: 10.1080/15384101.2020.1731946. Epub 2020 Feb 27. Cell Cycle. 2020. Retraction in: Cell Cycle. 2023 Mar-Mar;22(6):736. doi: 10.1080/15384101.2023.2171196. PMID: 32106756 Free PMC article. Retracted.
-
Exosomal long noncoding RNA MLETA1 promotes tumor progression and metastasis by regulating the miR-186-5p/EGFR and miR-497-5p/IGF1R axes in non-small cell lung cancer.J Exp Clin Cancer Res. 2023 Oct 26;42(1):283. doi: 10.1186/s13046-023-02859-y. J Exp Clin Cancer Res. 2023. PMID: 37880793 Free PMC article.
-
MicroRNA-92a promotes epithelial-mesenchymal transition through activation of PTEN/PI3K/AKT signaling pathway in non-small cell lung cancer metastasis.Int J Oncol. 2017 Jul;51(1):235-244. doi: 10.3892/ijo.2017.3999. Epub 2017 May 16. Int J Oncol. 2017. PMID: 28534966
-
miR-26a is a Key Therapeutic Target with Enormous Potential in the Diagnosis and Prognosis of Human Disease.Curr Med Chem. 2024;31(18):2550-2570. doi: 10.2174/0109298673271808231116075056. Curr Med Chem. 2024. PMID: 38204224 Review.
Cited by
-
The Role of lncRNA-miR-26a-mRNA Network in Cancer Progression and Treatment.Biochem Genet. 2024 Jun;62(3):1443-1461. doi: 10.1007/s10528-023-10475-w. Epub 2023 Sep 20. Biochem Genet. 2024. PMID: 37730965 Review.
-
Pancreatic cancer cells hijack tumor suppressive microRNA-26a to promote radioresistance and potentiate tumor repopulation.Heliyon. 2024 May 15;10(10):e31346. doi: 10.1016/j.heliyon.2024.e31346. eCollection 2024 May 30. Heliyon. 2024. PMID: 38807872 Free PMC article.
-
A Genomic Instability-Related Long Noncoding RNA Signature for Predicting Hepatocellular Carcinoma Prognosis.J Oncol. 2022 Aug 29;2022:3090523. doi: 10.1155/2022/3090523. eCollection 2022. J Oncol. 2022. PMID: 36072979 Free PMC article.
-
Oxidative Stress and Inflammation in Cardiovascular Diseases and Cancer: Role of Non-coding RNAs.Yale J Biol Med. 2022 Mar 31;95(1):129-152. eCollection 2022 Mar. Yale J Biol Med. 2022. PMID: 35370493 Free PMC article. Review.
-
MiRNA-423 rs6505162 and miRNA-6811 rs2292879 SNP associated with lung cancer in Hainan, China.Biosci Rep. 2023 Sep 27;43(9):BSR20231152. doi: 10.1042/BSR20231152. Biosci Rep. 2023. PMID: 37694278 Free PMC article.
References
-
- Zheng S-R, Sun K-X, Zhang S-W, Zeng H-M, Zou X-N, Chen R. Analysis of cancer prevalence in China in 2015. Chin J Oncol. 2019;41:19–28. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

