The lncRNA RUNX1-IT1 regulates C-FOS transcription by interacting with RUNX1 in the process of pancreatic cancer proliferation, migration and invasion
- PMID: 32487998
- PMCID: PMC7265432
- DOI: 10.1038/s41419-020-2617-7
The lncRNA RUNX1-IT1 regulates C-FOS transcription by interacting with RUNX1 in the process of pancreatic cancer proliferation, migration and invasion
Abstract
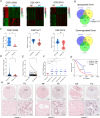
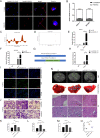
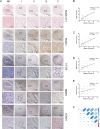
Numerous long noncoding RNAs (lncRNAs) are aberrantly expressed in pancreatic cancer (PC); however, their functions and mechanisms in cancer progression are largely unknown. In this study, we identified a novel PC-associated lncRNA, RUNX1-IT1, that was significantly upregulated in PC patient samples from multiple centers and associated with poor prognosis. In vitro and in vivo, alterations in RUNX1-IT1 expression markedly affected PC proliferation, migration and invasion. RUNX1-IT1 contributed to the progression of PC by interacting with the adjacent gene RUNX1. Rescue experiments showed that RUNX1 reduced the cancer-promoting effect of RUNX1-IT1. RNA-seq analysis after silencing RUNX1-IT1 and RUNX1 highlighted alterations in the common target C-FOS. Mechanistically, we demonstrated that RUNX1-IT1 was a trans-acting factor that participated in the proliferation, migration and invasion of PC by recruiting RUNX1 to the C-FOS gene promoter. Furthermore, RUNX1-IT1 enhanced the transcription of the RUNX1 gene, indicating its potential as a cis-regulatory RNA involved in the upstream regulation of RUNX1. Overall, RUNX1-IT1 is a crucial oncogenic lncRNA that activates C-FOS expression by regulating and recruiting RUNX1 and is a potential prognostic biomarker and therapeutic target for PC.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Similar articles
-
LncRNA RUNX1-IT1 which is downregulated by hypoxia-driven histone deacetylase 3 represses proliferation and cancer stem-like properties in hepatocellular carcinoma cells.Cell Death Dis. 2020 Feb 5;11(2):95. doi: 10.1038/s41419-020-2274-x. Cell Death Dis. 2020. PMID: 32024815 Free PMC article.
-
Long noncoding RNA CPS1-IT1 suppresses the metastasis of hepatocellular carcinoma by regulating HIF-1α activity and inhibiting epithelial-mesenchymal transition.Oncotarget. 2016 Jul 12;7(28):43588-43603. doi: 10.18632/oncotarget.9635. Oncotarget. 2016. PMID: 27248828 Free PMC article.
-
Long non-coding RNA RUNX1-IT1 plays a tumour-suppressive role in colorectal cancer by inhibiting cell proliferation and migration.Cell Biochem Funct. 2019 Jan;37(1):11-20. doi: 10.1002/cbf.3368. Epub 2018 Nov 30. Cell Biochem Funct. 2019. PMID: 30499136
-
LncRNAs in pancreatic cancer.Oncotarget. 2016 Aug 30;7(35):57379-57390. doi: 10.18632/oncotarget.10545. Oncotarget. 2016. PMID: 27429196 Free PMC article. Review.
-
LINC00473: A novel oncogenic long noncoding RNA in human cancers.J Cell Physiol. 2021 Jun;236(6):4174-4183. doi: 10.1002/jcp.30176. Epub 2020 Nov 22. J Cell Physiol. 2021. PMID: 33222224 Review.
Cited by
-
Regulatory function and mechanism research for m6A modification WTAP via SUCLG2-AS1- miR-17-5p-JAK1 axis in AML.BMC Cancer. 2024 Jan 17;24(1):98. doi: 10.1186/s12885-023-11687-4. BMC Cancer. 2024. PMID: 38233760 Free PMC article.
-
Gene expression profiles and bioinformatics analysis in lung samples from ovalbumin-induced asthmatic mice.BMC Pulm Med. 2023 Feb 2;23(1):50. doi: 10.1186/s12890-023-02306-w. BMC Pulm Med. 2023. PMID: 36726128 Free PMC article.
-
RUNX1-IT1 favors breast cancer carcinogenesis through regulation of IGF2BP1/GPX4 axis.Discov Oncol. 2023 Apr 10;14(1):42. doi: 10.1007/s12672-023-00652-z. Discov Oncol. 2023. PMID: 37036576 Free PMC article.
-
Long non‑coding RNA MIR4435‑2HG promotes the progression of head and neck squamous cell carcinoma by regulating the miR‑383‑5p/RBM3 axis.Oncol Rep. 2021 Jun;45(6):99. doi: 10.3892/or.2021.8050. Epub 2021 Apr 13. Oncol Rep. 2021. PMID: 33846802 Free PMC article.
-
Exploring the Regulatory Role of ncRNA in NAFLD: A Particular Focus on PPARs.Cells. 2022 Dec 7;11(24):3959. doi: 10.3390/cells11243959. Cells. 2022. PMID: 36552725 Free PMC article. Review.
References
-
- Habib J, Yu J. Circulating tumor cells in pancreatic cancer. J. Pancreatol. 2019;2:54–59. doi: 10.1097/JP9.0000000000000014. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

