Dosage effect of multiple genes accounts for multisystem disorder of myotonic dystrophy type 1
- PMID: 31853004
- PMCID: PMC7015062
- DOI: 10.1038/s41422-019-0264-2
Dosage effect of multiple genes accounts for multisystem disorder of myotonic dystrophy type 1
Abstract
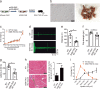
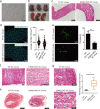
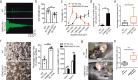
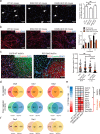
Multisystem manifestations in myotonic dystrophy type 1 (DM1) may be due to dosage reduction in multiple genes induced by aberrant expansion of CTG repeats in DMPK, including DMPK, its neighboring genes (SIX5 or DMWD) and downstream MBNL1. However, direct evidence is lacking. Here, we develop a new strategy to generate mice carrying multigene heterozygous mutations to mimic dosage reduction in one step by injection of haploid embryonic stem cells with mutant Dmpk, Six5 and Mbnl1 into oocytes. The triple heterozygous mutant mice exhibit adult-onset DM1 phenotypes. With the additional mutation in Dmwd, the quadruple heterozygous mutant mice recapitulate many major manifestations in congenital DM1. Moreover, muscle stem cells in both models display reduced stemness, providing a unique model for screening small molecules for treatment of DM1. Our results suggest that the complex symptoms of DM1 result from the reduced dosage of multiple genes.
Conflict of interest statement
The authors declare no competing interests.
Figures

Comment in
-
New myotonic dystrophy type 1 mouse model.Cell Res. 2020 Feb;30(2):99-100. doi: 10.1038/s41422-020-0276-y. Cell Res. 2020. PMID: 31953529 Free PMC article. No abstract available.
Similar articles
-
Sense and Antisense DMPK RNA Foci Accumulate in DM1 Tissues during Development.PLoS One. 2015 Sep 4;10(9):e0137620. doi: 10.1371/journal.pone.0137620. eCollection 2015. PLoS One. 2015. PMID: 26339785 Free PMC article.
-
Fuchs' Endothelial Corneal Dystrophy and RNA Foci in Patients With Myotonic Dystrophy.Invest Ophthalmol Vis Sci. 2017 Sep 1;58(11):4579-4585. doi: 10.1167/iovs.17-22350. Invest Ophthalmol Vis Sci. 2017. PMID: 28886202 Free PMC article.
-
Epigenetics of the myotonic dystrophy-associated DMPK gene neighborhood.Epigenomics. 2016 Jan;8(1):13-31. doi: 10.2217/epi.15.104. Epub 2016 Jan 12. Epigenomics. 2016. PMID: 26756355 Free PMC article.
-
Myotonic dystrophy: clinical and molecular parallels between myotonic dystrophy type 1 and type 2.Curr Neurol Neurosci Rep. 2002 Sep;2(5):465-70. doi: 10.1007/s11910-002-0074-6. Curr Neurol Neurosci Rep. 2002. PMID: 12169228 Review.
-
Myotonic dystrophy type 1: role of CCG, CTC and CGG interruptions within DMPK alleles in the pathogenesis and molecular diagnosis.Clin Genet. 2017 Oct;92(4):355-364. doi: 10.1111/cge.12954. Epub 2017 Feb 22. Clin Genet. 2017. PMID: 27991661 Review.
Cited by
-
RNA m6A methylation promotes the formation of vasculogenic mimicry in hepatocellular carcinoma via Hippo pathway.Angiogenesis. 2021 Feb;24(1):83-96. doi: 10.1007/s10456-020-09744-8. Epub 2020 Sep 13. Angiogenesis. 2021. PMID: 32920668
-
Medical care of rare and undiagnosed diseases: Prospects and challenges.Fundam Res. 2022 Sep 7;2(6):851-858. doi: 10.1016/j.fmre.2022.08.018. eCollection 2022 Nov. Fundam Res. 2022. PMID: 38933390 Free PMC article. Review.
-
Sequence composition changes in short tandem repeats: heterogeneity, detection, mechanisms and clinical implications.Nat Rev Genet. 2024 Jul;25(7):476-499. doi: 10.1038/s41576-024-00696-z. Epub 2024 Mar 11. Nat Rev Genet. 2024. PMID: 38467784 Review.
-
The pathogenesis of common Gjb2 mutations associated with human hereditary deafness in mice.Cell Mol Life Sci. 2023 May 13;80(6):148. doi: 10.1007/s00018-023-04794-9. Cell Mol Life Sci. 2023. PMID: 37178259 Free PMC article.
-
Calcitriol increases MBNL1 expression and alleviates myotonic dystrophy phenotypes in HSALR mouse models.J Transl Med. 2022 Dec 12;20(1):588. doi: 10.1186/s12967-022-03806-9. J Transl Med. 2022. PMID: 36510245 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

