The novel long noncoding RNA Lnc19959.2 modulates triglyceride metabolism-associated genes through the interaction with Purb and hnRNPA2B1
- PMID: 32302712
- PMCID: PMC7262451
- DOI: 10.1016/j.molmet.2020.100996
The novel long noncoding RNA Lnc19959.2 modulates triglyceride metabolism-associated genes through the interaction with Purb and hnRNPA2B1
Abstract
Objective: Long noncoding RNAs (lncRNAs) are currently considered to have a vital and wide range of biological functions, but the molecular mechanism underlying triglycerides metabolism remains poorly understood. This study aims to identify novel lncRNAs differentially expressed in rat livers with hypertriglyceridemia and elucidated the function role in TG metabolism.
Methods: Differentially expressions of lncRNAs in rat livers with hypertriglyceridemia were identified by transcriptome sequencing and validated by real-time PCR. The role of lnc19959.2 in triglyceride metabolism was assessed both in vitro and in vivo. RNA pulldown and RIP assays were conducted to evaluate the interactions between lnc19959.2 and its target proteins. ChIP and Dual report assays were performed to detect the interactions between transcription factors and promoters of its target genes.
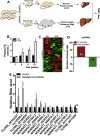
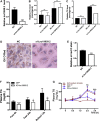
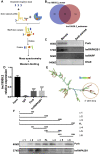
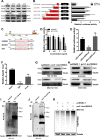
Results: We identified a novel lncRNA, and lnc19959.2 was upregulated in rat livers with hypertriglyceridemia. The knockdown of lnc19959.2 has profound TG lowering effects in vitro and in vivo. Subsequently, the genome-wide analysis identified that the knockdown of lnc19959.2 caused the deregulation of many genes during TG homeostasis. Further mechanism studies revealed that lnc19959.2 upregulated ApoA4 expression via ubiquitinated transcription inhibitor factor Purb, while it specifically interacted with hnRNPA2B1 to downregulate the expression of Cpt1a, Tm7sf2, and Gpam, respectively. In the upstream pathway, palmitate acid upregulated CCAAT/Enhancer-Binding Protein Beta (Cebpb) and facilitated its binding to the promoter of lnc19959.2, which resulted in significant promotion of lnc19959.2 transcriptional activity.
Conclusions: Our findings provide novel insights into a new layer regulatory complexity of an lncRNA modulating triglyceride homeostasis by a novel lncRNA lnc19959.2.
Keywords: Epigenetics; LncRNAs; Transcriptional regulation; Triglyceride metabolism.
Copyright © 2020 The Authors. Published by Elsevier GmbH.. All rights reserved.
Figures




Similar articles
-
Transcriptome Profile Analysis Reveals an Estrogen Induced LncRNA Associated with Lipid Metabolism and Carcass Traits in Chickens (Gallus Gallus).Cell Physiol Biochem. 2018;50(5):1638-1658. doi: 10.1159/000494785. Epub 2018 Nov 1. Cell Physiol Biochem. 2018. PMID: 30384372
-
Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma.Theranostics. 2018 Apr 15;8(10):2814-2829. doi: 10.7150/thno.23012. eCollection 2018. Theranostics. 2018. PMID: 29774077 Free PMC article.
-
A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism.Hepatology. 2016 Jul;64(1):58-72. doi: 10.1002/hep.28391. Epub 2016 Jan 22. Hepatology. 2016. PMID: 26663205
-
Long noncoding RNAs in lipid metabolism.Curr Opin Lipidol. 2018 Jun;29(3):224-232. doi: 10.1097/MOL.0000000000000503. Curr Opin Lipidol. 2018. PMID: 29553997 Free PMC article. Review.
-
Long noncoding RNAs: Novel insights into hepatocelluar carcinoma.Cancer Lett. 2014 Mar 1;344(1):20-27. doi: 10.1016/j.canlet.2013.10.021. Epub 2013 Oct 30. Cancer Lett. 2014. PMID: 24183851 Review.
Cited by
-
Long Noncoding RNAs: Novel Important Players in Adipocyte Lipid Metabolism and Derivative Diseases.Front Physiol. 2021 Jun 8;12:691824. doi: 10.3389/fphys.2021.691824. eCollection 2021. Front Physiol. 2021. PMID: 34168572 Free PMC article. Review.
-
APOA1/C3/A4/A5 Gene Cluster at 11q23.3 and Lipid Metabolism Disorders: From Epigenetic Mechanisms to Clinical Practices.Biomedicines. 2024 May 31;12(6):1224. doi: 10.3390/biomedicines12061224. Biomedicines. 2024. PMID: 38927431 Free PMC article. Review.
-
Role of long non-coding RNAs in adipogenesis: State of the art and implications in obesity and obesity-associated diseases.Obes Rev. 2021 Jul;22(7):e13203. doi: 10.1111/obr.13203. Epub 2021 Jan 14. Obes Rev. 2021. PMID: 33443301 Free PMC article. Review.
-
Transcriptome-metabolome analysis reveals how sires affect meat quality in hybrid sheep populations.Front Nutr. 2022 Aug 11;9:967985. doi: 10.3389/fnut.2022.967985. eCollection 2022. Front Nutr. 2022. PMID: 36034900 Free PMC article.
-
Long non-coding RNAs: a valuable biomarker for metabolic syndrome.Mol Genet Genomics. 2022 Sep;297(5):1169-1183. doi: 10.1007/s00438-022-01922-1. Epub 2022 Jul 19. Mol Genet Genomics. 2022. PMID: 35854006 Review.
References
-
- Nordestgaard B.G. Triglyceride-rich lipoproteins and atherosclerotic cardiovascular disease: new insights from epidemiology, genetics, and biology. Circulation Research. 2016;118(4):547–563. - PubMed
-
- Nordestgaard B.G., Varbo A. Triglycerides and cardiovascular disease. Lancet. 2014;384(9943):626–635. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

