Sulforaphene inhibits esophageal cancer progression via suppressing SCD and CDH3 expression, and activating the GADD45B-MAP2K3-p38-p53 feedback loop
- PMID: 32873775
- PMCID: PMC7463232
- DOI: 10.1038/s41419-020-02859-2
Sulforaphene inhibits esophageal cancer progression via suppressing SCD and CDH3 expression, and activating the GADD45B-MAP2K3-p38-p53 feedback loop
Abstract
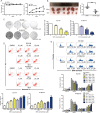
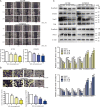
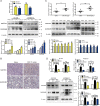
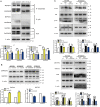
Esophageal cancer is one of the most common cancer with limited therapeutic strategies, thus it is important to develop more effective strategies to against it. Sulforaphene (SFE), an isothiocyanate isolated from radish seeds, was proved to inhibit esophageal cancer progression in the current study. Flow cytometric analysis showed SFE induced cell apoptosis and cycle arrest in G2/M phase. Also, scrape motility and transwell assays presented SFE reduced esophageal cancer cell metastasis. Microarray results showed the influence of SFE on esophageal cancer cells was related with stearoyl-CoA desaturase (SCD), cadherin 3 (CDH3), mitogen-activated protein kinase kinase 3 (MAP2K3) and growth arrest and DNA damage inducible beta (GADD45B). SCD and CDH3 could promote esophageal cancer metastasis via activating the Wnt pathway, while the latter one was involved in a positive feedback loop, GADD45B-MAP2K3-p38-p53, to suppress esophageal cancer growth. GADD45B was known to be the target gene of p53, and we proved in this study, it could increase the phosphorylation level of MAP2K3 in esophageal cancer cells, activating p38 and p53 in turn. SFE treatment elevated MAP2K3 and GADD45B expression and further stimulated this feedback loop to better exert antitumor effect. In summary, these results demonstrated that SFE had the potential for developing as a chemotherapeutic agent because of its inhibitory effects on esophageal cancer metastasis and proliferation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Sulforaphene inhibits triple negative breast cancer through activating tumor suppressor Egr1.Breast Cancer Res Treat. 2016 Jul;158(2):277-86. doi: 10.1007/s10549-016-3888-7. Epub 2016 Jul 4. Breast Cancer Res Treat. 2016. PMID: 27377973
-
Sulforaphene suppresses growth of colon cancer-derived tumors via induction of glutathione depletion and microtubule depolymerization.Mol Nutr Food Res. 2016 May;60(5):1068-78. doi: 10.1002/mnfr.201501011. Epub 2016 Mar 24. Mol Nutr Food Res. 2016. PMID: 26918318
-
The miR-19b-3p-MAP2K3-STAT3 feedback loop regulates cell proliferation and invasion in esophageal squamous cell carcinoma.Mol Oncol. 2021 May;15(5):1566-1583. doi: 10.1002/1878-0261.12934. Epub 2021 Mar 14. Mol Oncol. 2021. PMID: 33660414 Free PMC article.
-
Oridonin-induced mitochondria-dependent apoptosis in esophageal cancer cells by inhibiting PI3K/AKT/mTOR and Ras/Raf pathways.J Cell Biochem. 2019 Mar;120(3):3736-3746. doi: 10.1002/jcb.27654. Epub 2018 Sep 19. J Cell Biochem. 2019. PMID: 30229997
-
The natural compound sulforaphene, as a novel anticancer reagent, targeting PI3K-AKT signaling pathway in lung cancer.Oncotarget. 2016 Nov 22;7(47):76656-76666. doi: 10.18632/oncotarget.12307. Oncotarget. 2016. PMID: 27765931 Free PMC article.
Cited by
-
Development of a TLR-Based Model That Can Predict Prognosis, Tumor Microenvironment, and Drug Response for Esophageal Squamous Cell Carcinoma.Biochem Genet. 2024 Oct;62(5):3740-3760. doi: 10.1007/s10528-023-10629-w. Epub 2024 Jan 11. Biochem Genet. 2024. PMID: 38206423
-
Long noncoding RNAs with peptide-encoding potential identified in esophageal squamous cell carcinoma: KDM4A-AS1-encoded peptide weakens cancer cell viability and migratory capacity.Mol Oncol. 2023 Jul;17(7):1419-1436. doi: 10.1002/1878-0261.13424. Epub 2023 Apr 10. Mol Oncol. 2023. PMID: 36965032 Free PMC article.
-
Dissection of the MKK3 Functions in Human Cancer: A Double-Edged Sword?Cancers (Basel). 2022 Jan 18;14(3):483. doi: 10.3390/cancers14030483. Cancers (Basel). 2022. PMID: 35158751 Free PMC article. Review.
-
CRCDB: A comprehensive database for integrating and analyzing multi-omics data of early-onset and late-onset colorectal cancer.Comput Struct Biotechnol J. 2024 Jun 2;23:2507-2515. doi: 10.1016/j.csbj.2024.05.051. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38974887 Free PMC article.
-
Identifying the p65-Dependent Effect of Sulforaphene on Esophageal Squamous Cell Carcinoma Progression via Bioinformatics Analysis.Int J Mol Sci. 2020 Dec 23;22(1):60. doi: 10.3390/ijms22010060. Int J Mol Sci. 2020. PMID: 33374641 Free PMC article.
References
-
- Tseng CC, et al. Metabolic engineering probiotic yeast produces 3S, 3’S-astaxanthin to inhibit B16F10 metastasis. Food Chem. Toxicol. 2020;135:110993–111001. - PubMed
-
- Kuang PQ, et al. Preparative separation and purification of sulforaphene from radish seeds by high-speed countercurrent chromatography. Food Chem. 2013;136:342–347. - PubMed
-
- Kuang PQ, et al. Separation and purification of sulforaphene from radish seeds using macroporous resin and preparative high-performance liquid chromatography. Food Chem. 2013;136:342–347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

