Oplr16 serves as a novel chromatin factor to control stem cell fate by modulating pluripotency-specific chromosomal looping and TET2-mediated DNA demethylation
- PMID: 32055844
- PMCID: PMC7144914
- DOI: 10.1093/nar/gkaa097
Oplr16 serves as a novel chromatin factor to control stem cell fate by modulating pluripotency-specific chromosomal looping and TET2-mediated DNA demethylation
Abstract
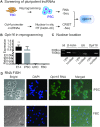
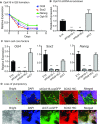
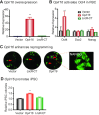
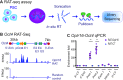
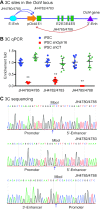
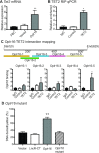
Formation of a pluripotency-specific chromatin network is a critical event in reprogramming somatic cells into pluripotent status. To characterize the regulatory components in this process, we used 'chromatin RNA in situ reverse transcription sequencing' (CRIST-seq) to profile RNA components that interact with the pluripotency master gene Oct4. Using this approach, we identified a novel nuclear lncRNA Oplr16 that was closely involved in the initiation of reprogramming. Oplr16 not only interacted with the Oct4 promoter and regulated its activity, but it was also specifically activated during reprogramming to pluripotency. Active expression of Oplr16 was required for optimal maintenance of pluripotency in embryonic stem cells. Oplr16 was also able to enhance reprogramming of fibroblasts into pluripotent cells. RNA reverse transcription-associated trap sequencing (RAT-seq) indicated that Oplr16 interacted with multiple target genes related to stem cell self-renewal. Of note, Oplr16 utilized its 3'-fragment to recruit the chromatin factor SMC1 to orchestrate pluripotency-specific intrachromosomal looping. After binding to the Oct4 promoter, Oplr16 recruited TET2 to induce DNA demethylation and activate Oct4 in fibroblasts, leading to enhanced reprogramming. These data suggest that Oplr16 may act as a pivotal chromatin factor to control stem cell fate by modulating chromatin architecture and DNA demethylation.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
LncRNA Osilr9 coordinates promoter DNA demethylation and the intrachromosomal loop structure required for maintaining stem cell pluripotency.Mol Ther. 2023 Jun 7;31(6):1791-1806. doi: 10.1016/j.ymthe.2022.12.010. Epub 2022 Dec 15. Mol Ther. 2023. PMID: 36523163 Free PMC article.
-
The roles of long noncoding RNAs in the regulation of OCT4 expression.Stem Cell Res Ther. 2022 Jul 30;13(1):383. doi: 10.1186/s13287-022-03059-9. Stem Cell Res Ther. 2022. PMID: 35907897 Free PMC article. Review.
-
Chromatin lncRNA Platr10 controls stem cell pluripotency by coordinating an intrachromosomal regulatory network.Genome Biol. 2021 Aug 19;22(1):233. doi: 10.1186/s13059-021-02444-6. Genome Biol. 2021. PMID: 34412677 Free PMC article.
-
Osblr8 orchestrates intrachromosomal loop structure required for maintaining stem cell pluripotency.Int J Biol Sci. 2020 Apr 6;16(11):1861-1875. doi: 10.7150/ijbs.45112. eCollection 2020. Int J Biol Sci. 2020. PMID: 32398955 Free PMC article.
-
Going up the hill: chromatin-based barriers to epigenetic reprogramming.FEBS J. 2021 Aug;288(16):4798-4811. doi: 10.1111/febs.15628. Epub 2020 Dec 6. FEBS J. 2021. PMID: 33190371 Review.
Cited by
-
Chromatin-RNA in situ Reverse Transcription Sequencing (CRIST-seq) Approach to Profile the Non-coding RNA Interaction Network.Bio Protoc. 2023 Jul 20;13(14):e4718. doi: 10.21769/BioProtoc.4718. eCollection 2023 Jul 20. Bio Protoc. 2023. PMID: 37497457 Free PMC article.
-
Effects of SMC1A on immune microenvironment and cancer stem cells in colon adenocarcinoma.Cancer Med. 2023 Jun;12(11):12881-12895. doi: 10.1002/cam4.5891. Epub 2023 Apr 25. Cancer Med. 2023. PMID: 37096492 Free PMC article.
-
LncRNA Osilr9 coordinates promoter DNA demethylation and the intrachromosomal loop structure required for maintaining stem cell pluripotency.Mol Ther. 2023 Jun 7;31(6):1791-1806. doi: 10.1016/j.ymthe.2022.12.010. Epub 2022 Dec 15. Mol Ther. 2023. PMID: 36523163 Free PMC article.
-
The roles of long noncoding RNAs in the regulation of OCT4 expression.Stem Cell Res Ther. 2022 Jul 30;13(1):383. doi: 10.1186/s13287-022-03059-9. Stem Cell Res Ther. 2022. PMID: 35907897 Free PMC article. Review.
-
Profiling mitochondria-polyribosome lncRNAs associated with pluripotency.Sci Data. 2023 Nov 2;10(1):755. doi: 10.1038/s41597-023-02649-3. Sci Data. 2023. PMID: 37919270 Free PMC article.
References
-
- Takahashi K., Yamanaka S.. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006; 126:663–676. - PubMed
-
- Andrey G., Mundlos S.. The three-dimensional genome: regulating gene expression during pluripotency and development. Development. 2017; 144:3646–3658. - PubMed
-
- de Wit E., Bouwman B.A., Zhu Y., Klous P., Splinter E., Verstegen M.J., Krijger P.H., Festuccia N., Nora E.P., Welling M. et al. .. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013; 501:227–231. - PubMed
-
- Li M., Liu G.H., Izpisua Belmonte J.C.. Navigating the epigenetic landscape of pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 2012; 13:524–535. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

