Loss of SIRT4 promotes the self-renewal of Breast Cancer Stem Cells
- PMID: 32863939
- PMCID: PMC7449925
- DOI: 10.7150/thno.44688
Loss of SIRT4 promotes the self-renewal of Breast Cancer Stem Cells
Abstract
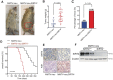
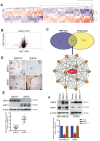
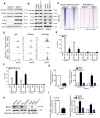
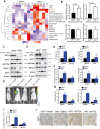
Rationale: It has been proposed that cancer stem/progenitor cells (or tumor-initiating cells, TICs) account for breast cancer initiation and progression. Sirtuins are nicotinamide adenine dinucleotide (NAD+)-dependent class-III histone deacetylases and mediate various basic biological processes, including metabolic homeostasis. However, interplay and cross-regulation among the sirtuin family are not fully understood. As one of the least studied sirtuin family members, the mitochondrial sirtuin SIRT4 is a tumor suppressor gene in various cancers. However, its role in cancer stemness, as well as initiation and progression of breast cancer, remains unknown. Methods: The expression of SIRT4 in breast cancer was analyzed using the TCGA breast cancer database and 3 GSEA data. Normal breast epithelial cells MCF10A and breast cancer cell lines MCF-7, MDA-MB-231, BT549, MDA-MB-468 were used to establish SIRT4 gene knockdown and corresponding overexpression cells. Identified MTT cytotoxicity assays, cell invasion and motility assay, sorting of SP, confocal immunofluorescence microscopy, mouse mammary stem cell analysis, glutamine and glucose production, clonogenic and sphere-formation assay, mass spectrometric metabolomics analysis and ChIP-seq to further explore SIRT4 biological role in breast cancer. Results: We elucidated a novel role for SIRT4 in the negative regulation of mammary gland development and stemness, which is related to the mammary tumorigenesis. We also uncovered an inverse correlation between SIRT4 and SIRT1. Most importantly, SIRT4 negatively regulates SIRT1 expression via repressing glutamine metabolism. Besides, we identified H4K16ac and BRCA1 as new prime targets of SIRT4 in breast cancer. Conclusions: These results demonstrate that SIRT4 exerts its tumor-suppressive activity via modulating SIRT1 expression in breast cancer and provide a novel cross-talk between mitochondrial and nuclear sirtuins.
Keywords: SIRT1; SIRT4; breast cancer; cancer stemness; glutamine metabolism.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




Similar articles
-
SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma.J Biol Chem. 2014 Feb 14;289(7):4135-44. doi: 10.1074/jbc.M113.525949. Epub 2013 Dec 24. J Biol Chem. 2014. PMID: 24368766 Free PMC article.
-
SIRT4 functions as a tumor suppressor during prostate cancer by inducing apoptosis and inhibiting glutamine metabolism.Sci Rep. 2022 Jul 16;12(1):12208. doi: 10.1038/s41598-022-16610-8. Sci Rep. 2022. PMID: 35842463 Free PMC article.
-
SIRT4 represses peroxisome proliferator-activated receptor α activity to suppress hepatic fat oxidation.Mol Cell Biol. 2013 Nov;33(22):4552-61. doi: 10.1128/MCB.00087-13. Epub 2013 Sep 16. Mol Cell Biol. 2013. PMID: 24043310 Free PMC article.
-
SIRT4 is the last puzzle of mitochondrial sirtuins.Bioorg Med Chem. 2018 Aug 7;26(14):3861-3865. doi: 10.1016/j.bmc.2018.07.031. Epub 2018 Jul 19. Bioorg Med Chem. 2018. PMID: 30033389 Review.
-
Mammalian Sirtuins SIRT4 and SIRT7.Prog Mol Biol Transl Sci. 2018;154:147-168. doi: 10.1016/bs.pmbts.2017.11.001. Epub 2018 Jan 19. Prog Mol Biol Transl Sci. 2018. PMID: 29413176 Review.
Cited by
-
Progress of Breast Cancer basic research in China.Int J Biol Sci. 2021 May 11;17(8):2069-2079. doi: 10.7150/ijbs.60631. eCollection 2021. Int J Biol Sci. 2021. PMID: 34131406 Free PMC article. Review.
-
Research Progress of Sirtuin4 in Cancer.Front Oncol. 2021 Jan 8;10:562950. doi: 10.3389/fonc.2020.562950. eCollection 2020. Front Oncol. 2021. PMID: 33585187 Free PMC article. Review.
-
CCL16 maintains stem cell-like properties in breast cancer by activating CCR2/GSK3β/β-catenin/OCT4 axis.Theranostics. 2021 Jan 1;11(5):2297-2317. doi: 10.7150/thno.51000. eCollection 2021. Theranostics. 2021. PMID: 33500726 Free PMC article.
-
Mitochondrial Sirtuins in Chronic Degenerative Diseases: New Metabolic Targets in Colorectal Cancer.Int J Mol Sci. 2022 Mar 16;23(6):3212. doi: 10.3390/ijms23063212. Int J Mol Sci. 2022. PMID: 35328633 Free PMC article. Review.
-
CDC45 promotes the stemness and metastasis in lung adenocarcinoma by affecting the cell cycle.J Transl Med. 2024 Apr 8;22(1):335. doi: 10.1186/s12967-024-05038-5. J Transl Med. 2024. PMID: 38589907 Free PMC article.
References
-
- Zhu S, Dong Z, Ke X, Hou J, Zhao E, Zhang K, The roles of sirtuins family in cell metabolism during tumor development. Semin Cancer Biol. 2018. - PubMed
-
- Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

