Hsa_circ_0060450 Negatively Regulates Type I Interferon-Induced Inflammation by Serving as miR-199a-5p Sponge in Type 1 Diabetes Mellitus
- PMID: 33133095
- PMCID: PMC7550460
- DOI: 10.3389/fimmu.2020.576903
Hsa_circ_0060450 Negatively Regulates Type I Interferon-Induced Inflammation by Serving as miR-199a-5p Sponge in Type 1 Diabetes Mellitus
Abstract
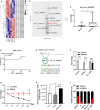
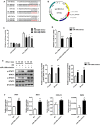
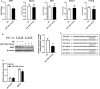
Circular RNAs (circRNAs) constitute a class of covalently circular non-coding RNA molecules formed by 5' and 3' end back-splicing. The rapid development of bioinformatics and large-scale sequencing has led to the identification of functional circRNAs. Despite an overall upward trend, studies focusing on the roles of circRNAs in immune diseases remain relatively scarce. In the present study, we obtained a differential circRNA expression profile based on microarray analysis of peripheral blood mononuclear cells (PBMCs) in children with type 1 diabetes mellitus (T1DM). We characterized one differentially expressed circRNA back-spliced from the MYB Proto-Oncogene Like 2 (MYBL2) gene in patients with T1DM, termed as hsa_circ_0060450. Subsequent assays revealed that hsa_circ_0060450 can serve as the sponge of miR-199a-5p, release its target gene, Src homology 2 (SH2)-containing protein tyrosine phosphatase 2 (SHP2), encoded by the tyrosine-protein phosphatase non-receptor type 11 gene (PTPN11), and further suppress the JAK-STAT signaling pathway triggered by type I interferon (IFN-I) to inhibit macrophage-mediated inflammation, which indicates the important roles of circRNAs in T1DM and represents a promising therapeutic molecule in the treatment of T1DM.
Keywords: SHP2; circular RNA; macrophage; type 1 diabetes mellitus; type I interferon.
Copyright © 2020 Yang, Han, Zhang, Sun, Huang, Xiao, Gao, Liang, Luo, Lu, Fu and Zhou.
Figures


Similar articles
-
Overexpression of circ_0021739 in Peripheral Blood Mononuclear Cells in Women with Postmenopausal Osteoporosis Is Associated with Reduced Expression of microRNA-194-5p in Osteoclasts.Med Sci Monit. 2021 Apr 20;27:e929170. doi: 10.12659/MSM.929170. Med Sci Monit. 2021. PMID: 33875631 Free PMC article.
-
Circular RNA circPPM1F modulates M1 macrophage activation and pancreatic islet inflammation in type 1 diabetes mellitus.Theranostics. 2020 Aug 29;10(24):10908-10924. doi: 10.7150/thno.48264. eCollection 2020. Theranostics. 2020. PMID: 33042261 Free PMC article.
-
Transcriptome sequencing of circular RNA reveals a novel circular RNA-has_circ_0114427 in the regulation of inflammation in acute kidney injury.Clin Sci (Lond). 2020 Jan 31;134(2):139-154. doi: 10.1042/CS20190990. Clin Sci (Lond). 2020. PMID: 31930399
-
Circular RNAs and inflammation: Epigenetic regulators with diagnostic role.Pathol Res Pract. 2023 Nov;251:154912. doi: 10.1016/j.prp.2023.154912. Epub 2023 Oct 26. Pathol Res Pract. 2023. PMID: 38238072 Review.
-
circGFRA1: A circular RNA with important roles in human carcinogenesis.Pathol Res Pract. 2023 Aug;248:154588. doi: 10.1016/j.prp.2023.154588. Epub 2023 Jun 1. Pathol Res Pract. 2023. PMID: 37285736 Review.
Cited by
-
Targeting protein phosphatases for the treatment of inflammation-related diseases: From signaling to therapy.Signal Transduct Target Ther. 2022 Jun 4;7(1):177. doi: 10.1038/s41392-022-01038-3. Signal Transduct Target Ther. 2022. PMID: 35665742 Free PMC article. Review.
-
Understanding Competitive Endogenous RNA Network Mechanism in Type 1 Diabetes Mellitus Using Computational and Bioinformatics Approaches.Diabetes Metab Syndr Obes. 2021 Sep 8;14:3865-3945. doi: 10.2147/DMSO.S315488. eCollection 2021. Diabetes Metab Syndr Obes. 2021. PMID: 34526791 Free PMC article.
-
Differential Expression and Bioinformatics Analysis of Plasma-Derived Exosomal circRNA in Type 1 Diabetes Mellitus.J Immunol Res. 2022 Oct 27;2022:3625052. doi: 10.1155/2022/3625052. eCollection 2022. J Immunol Res. 2022. PMID: 36339941 Free PMC article.
-
MicroRNA signatures in the pathogenesis and therapy of inflammatory bowel disease.Clin Exp Med. 2024 Sep 11;24(1):217. doi: 10.1007/s10238-024-01476-z. Clin Exp Med. 2024. PMID: 39259390 Free PMC article. Review.
-
The crucial regulatory role of type I interferon in inflammatory diseases.Cell Biosci. 2023 Dec 20;13(1):230. doi: 10.1186/s13578-023-01188-z. Cell Biosci. 2023. PMID: 38124132 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

